Welcome to the Walhout Lab
The Structure, Function and Evolution of Biological Networks
Metabolic networks provide building blocks and energy to support organism development, growth, wound healing, homeostasis, and response to nutritional, environmental and therapeutic inputs, therefore the tight regulation of these networks is vital to organismal fitness. In the Walhout lab we aim to understand how metabolic networks are organized and regulated, how their organization enables function, and how these networks evolve. We are particularly interested in the communication between metabolic and gene regulatory networks and ask important questions at both broad systems-level as well as deep mechanistic levels to understand these biological processes. Our model organism of choice is the roundworm Caenorhabditis elegans. These worms are highly adaptable, easy to manipulate, and have many analogs in human genetics. We have made significant advances to understand the composition and tissue specific functions of metabolic networks using various experimental and computational methods such as large-scale genetic screens, transcriptomics, metabolomics and transcription factor binding assays, as well as flux balance analysis, nested effects modeling and statistics. However, there is still more work to done. We welcome you to join us on our journey!
Latest Publications
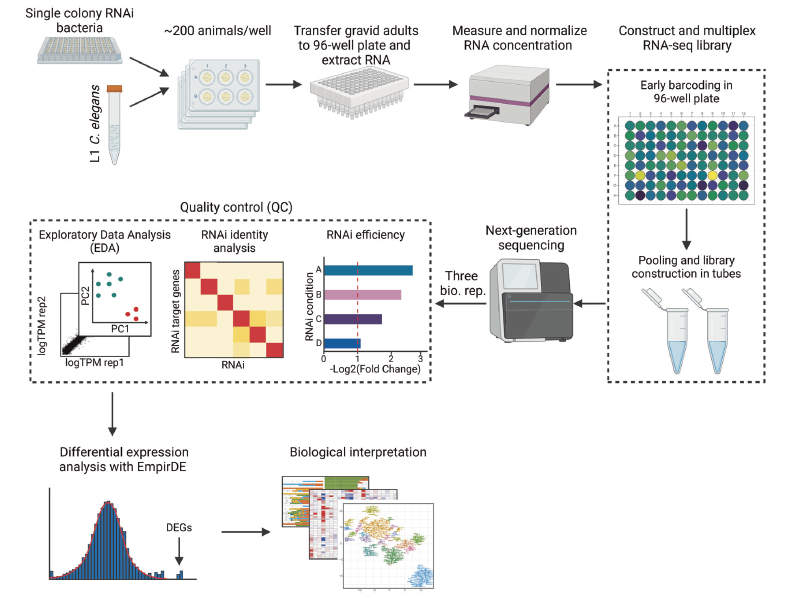
Worm Perturb-Seq: massively parallel whole-animal RNAi and RNA-seq
Zhang, Li et al.
Systems-level design principles of metabolic rewiring in an animal
Li, Zhang et al.
Above papers highlighted in Science Daily
A systems-level, semiquantitative landscape of metabolic flux in C. elegans
Zhang, Li et al.
Meet the Team